The antibiotic-resistant bacteria has emerged as a serious threat to public health. The World Health Organization (WHO) has developed a global priority pathogens list (global PPL) of antibiotic-resistant bacteria to help prioritizing the research and development (R&D) of new and effective antibiotic treatments. Our team aims to become a trustworthy partner who specialises on below subjects.
16S Metagenomics (MICROBIOME ANALYSIS)
Metagenomic studies using various samples (stool, blood, saliva, tissue, etc.)
16S rRNA targeted sequencing
Short-read next-generation sequencing (MiSeq System, illumina)
Long-read next-generation sequencing (MinION, Oxford Nanopore Technologies)
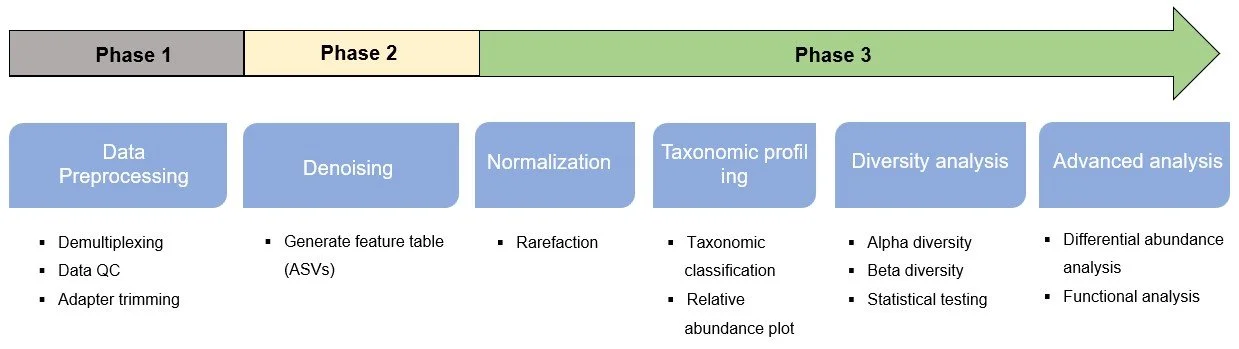
Data QC and Analysis
FastQC : fastq data qulity control
DADA2 : denoise reads into ASVs
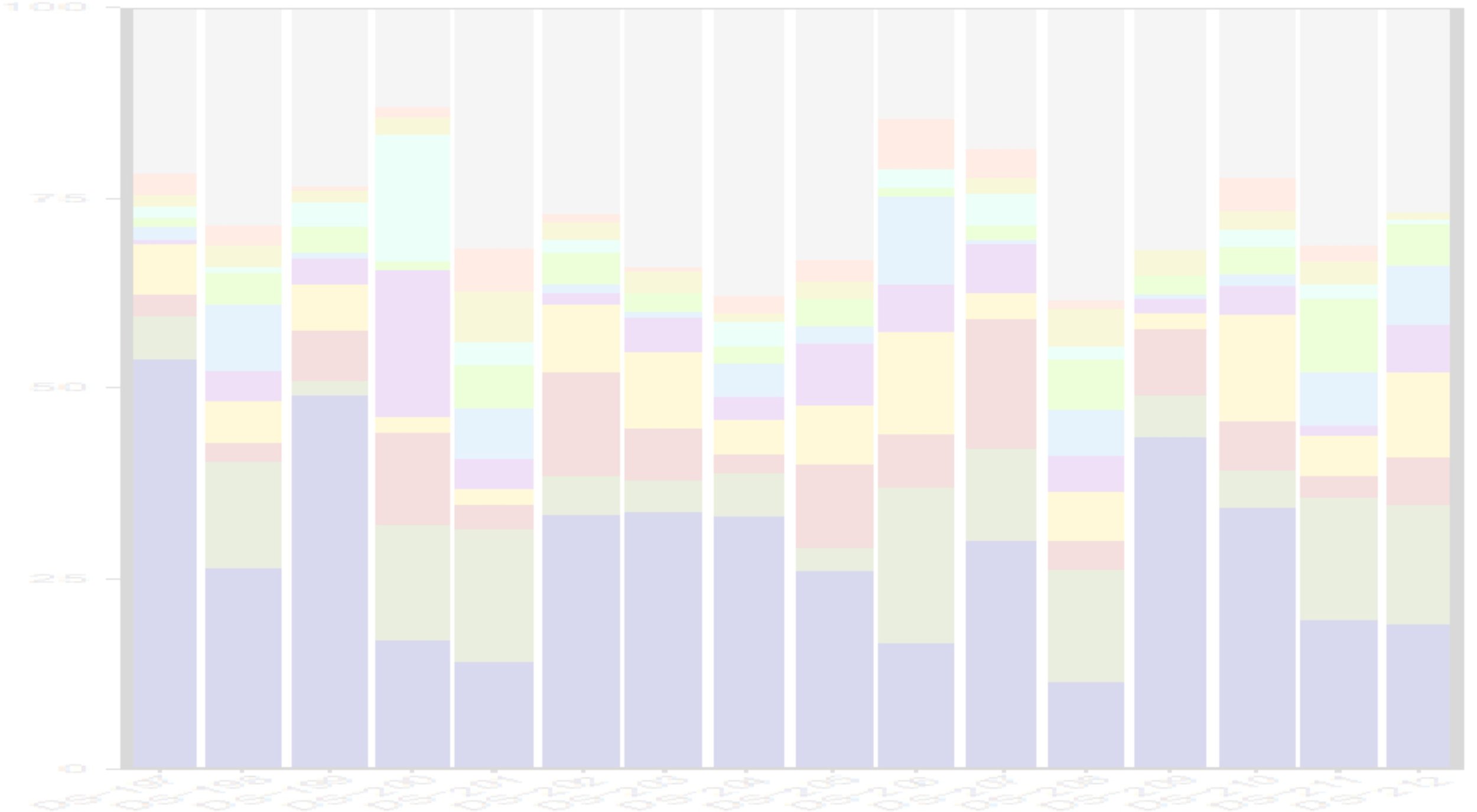
QIIME2 & R : taxonomic profiling, Diversity analysis, Abundance analysis
Sequence Database : GREENGENES, Silva, rdp
16S rRNA targeted sequencing analysis pipeline
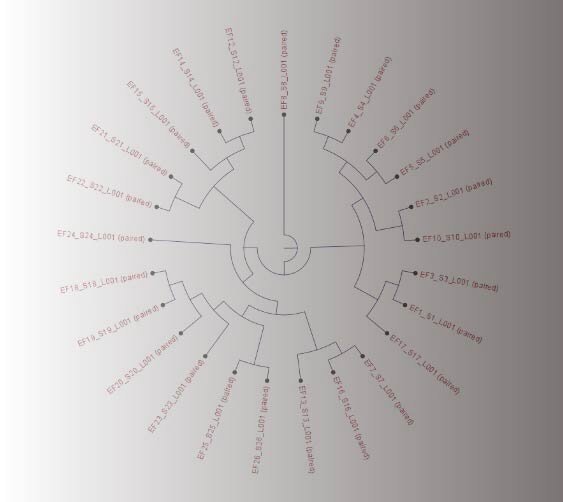
Epidemiological analysis
Whole-Genom Sequencing (WGS), Multilocus Sequence Typing (MLST) using next-generation sequencing
Short-read next-generation sequencing (MiSeq System, illumina)
Long-read next-generation sequencing (MinION, Oxford Nanopore Technologies)
Bacteria Identification
16S rRNA gene analysis by Sanger sequencing
Targetted bacteria detection by Real-time PCR
H. pylori, A. baumannii, Campylobacter, N. gonorrhoeae, S. pneumoniae, H. influenzae, Salmonella, Shigella, etc.
Anti-Microbial Resistance
Modular multiplex real-time PCR (MMRT-PCR)-based anti-microbial resistance gene detection
Carbapenem : Enterobacteriaceae, Pseudomonas aeruginosa, Acinetobacter baumannii
Cephalosporin : Enterobacteriacea, Neisseria gonorrhoeae
Vancomycin : Enterococcus faecium
Methicillin : Staphylococcus aureus
Clarithromycin : Helicobacter pylori
Fluoroquinolone : Campylobacter, Neisseria gonorrhoeae, Salmonella spp., Shigella spp.
Penicillin : Streptococcus pneumoniae
Ampicillin : Heamophilus influenzae